Transcriptional Factors Database
A bioinformatics project to develop an online database of MYB TFs in apple.
This project was proposed and done in group in The Basics of Bioinformation Technology (2022 Spring) taught by Prof. Jingchu Luo.
Enlightened by PlantTFDB, a holistic database of transcriptional factors (TFs) in plants, we are motivated to develop a smaller-scope database presenting information of MYB TFs in apple (Malus x domestica). It was available on the LEB course server.

Because the server is updated every academic year for new classes, the website wouldn’t be online forever. Please refer to this demo document if the link above is expired.
The GitHub repository is here.
Sequence alignment and data analysis
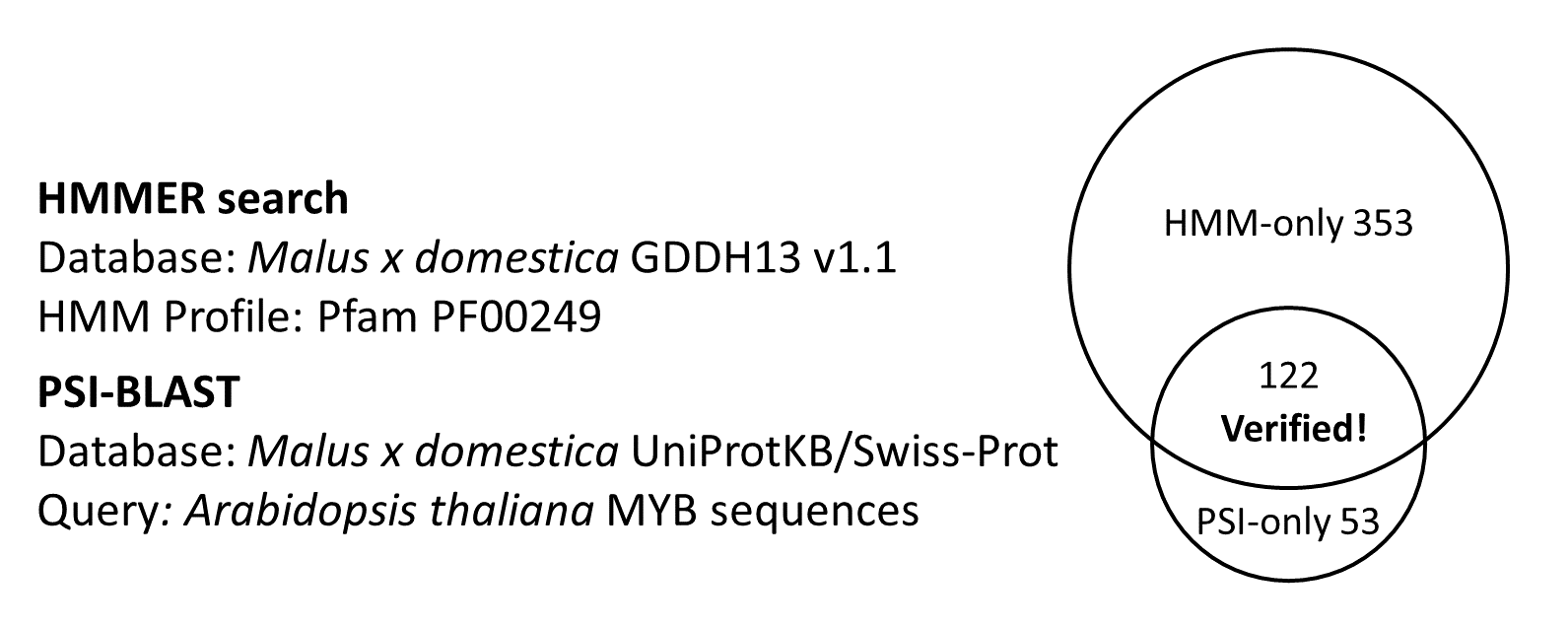
We selected Malus x Domestica GDDH13 v1.1 Whole Genome Assembly as reference sequence. Using HMMER search and PSI-BLAST in command line, we acquired 475 HMMER-identified sequences in which 122 of them were further verified by PSI-BLAST.
Database construction and visualization
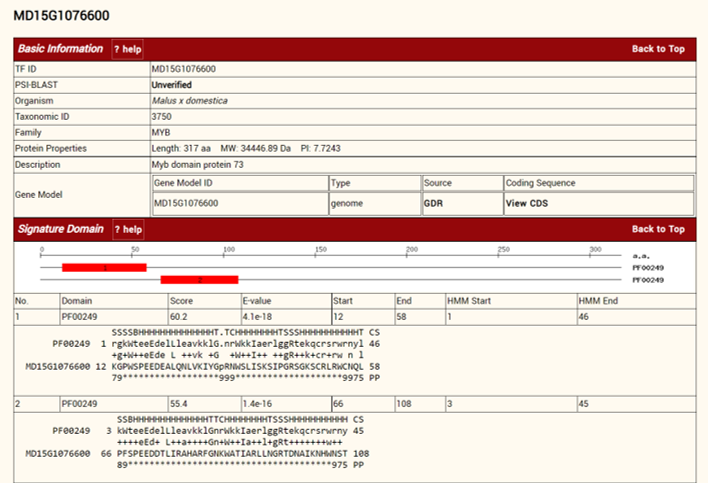
We then constructed a mySQL database and used PHP to build a web interface for the real-time visualization of the data. I built a php script to generate multiple pages for each TF, which can be accessed by clicking the TF name in the table. In each page, TF information, sequence alignment results were displayed.
Summary
This project was a great experience for me to practice bioinformatic algorithm and learn how to build a database from scratch. The experiences using multiple programming tools (i.e., command line, PHP, HTML, mySQL) simultaneously to analyze and present data are very interesting. It also gave me a better feeling of how a real-world bioinformatics project is done.
Reference
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., & Gao, G. (2016). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic acids research, gkw982.