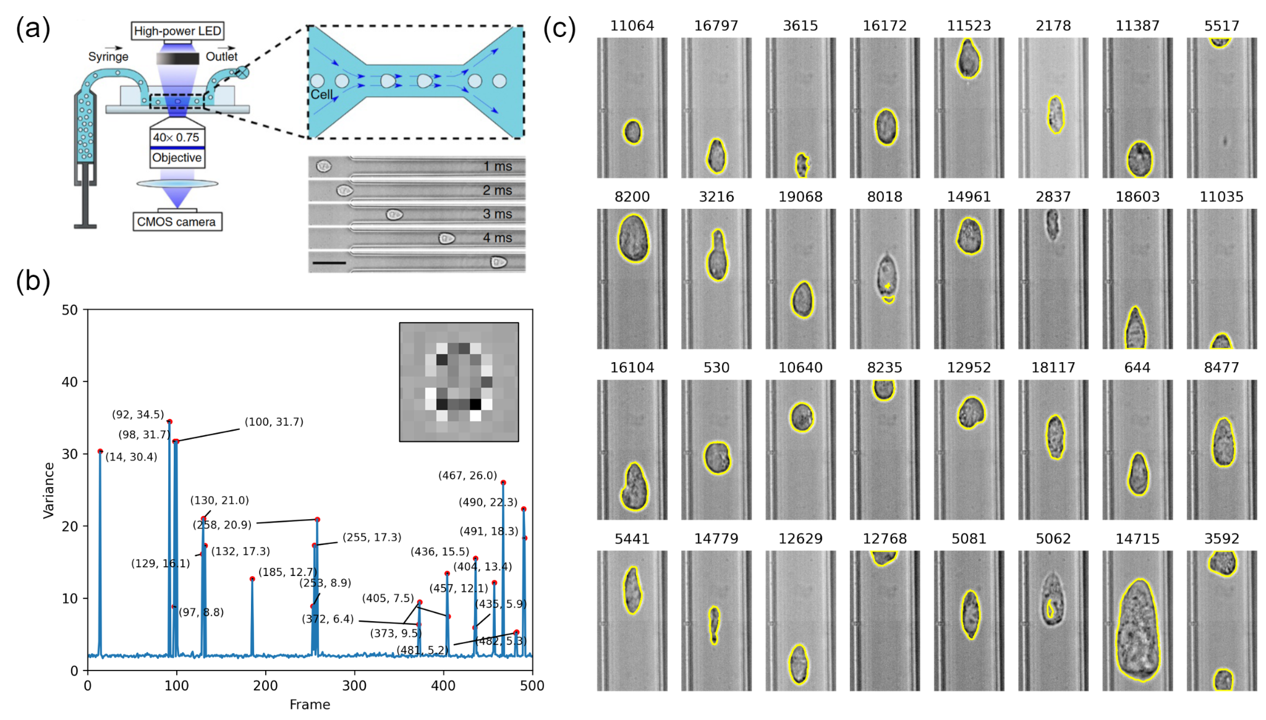
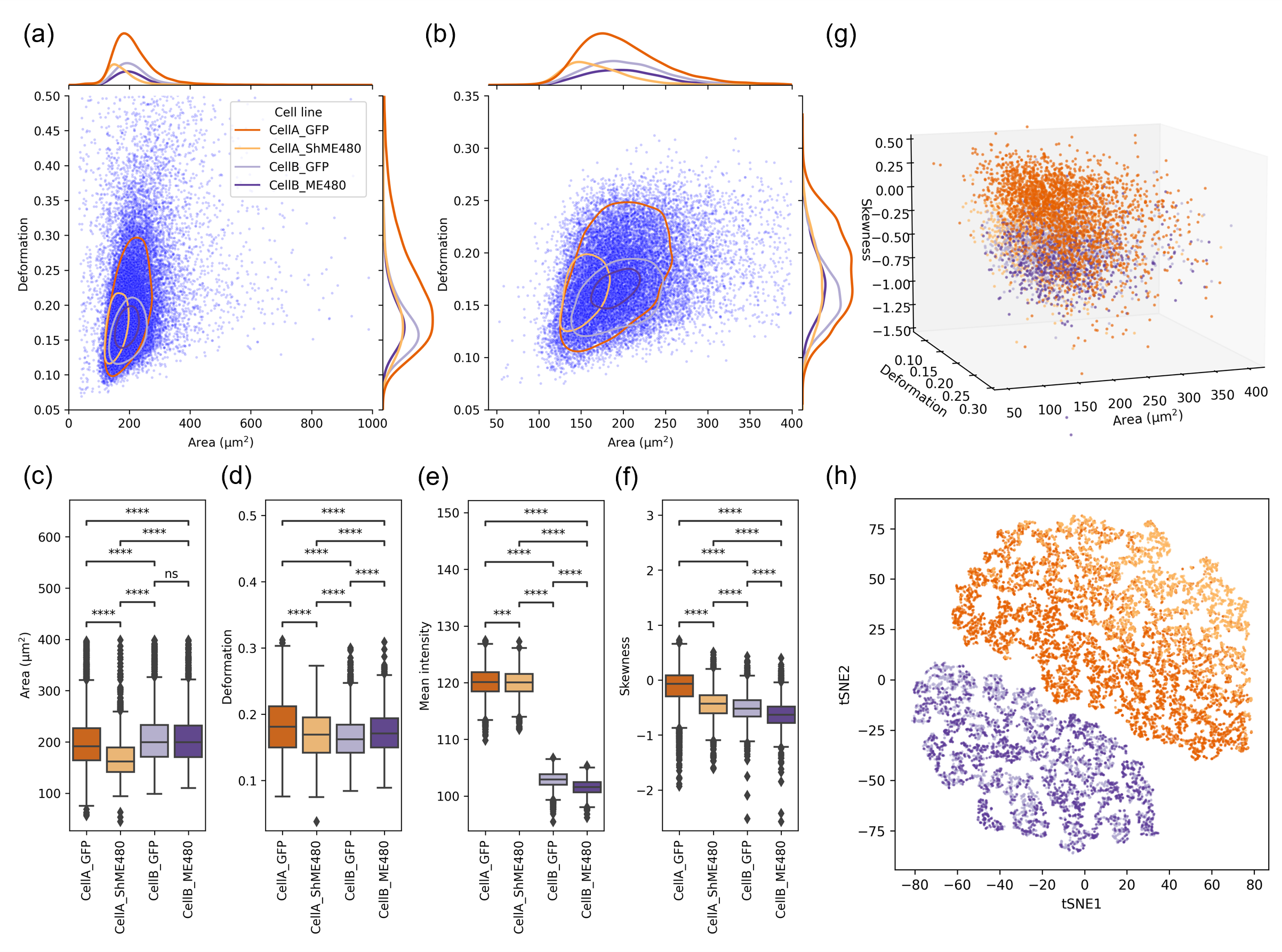
Deformation cytometry analysis
A bioimage processing workflow that segments and quantifies deformation cytometry data.
This project was done in group in Mechanobiology (2023 Fall) taught by Prof. Persat Alexandre and Prof. Sakar Selman.
Deformation cytometry (DC) is a flow-based technique which allows the high-throughput assessment of single-cell mechanical properties. It was proposed by Gossett. et al and further improved by Otto. et al. Within the microfluidic channel, cells are deformed by hydrodynamic constriction and their morphological changes are recorded by high-speed imaging. Here we implemented the analytical part of DC. Specifically, I focused on the development of image analysis workflow. Two major tasks were included:
- Image stack size reduction: DC raw data is a huge time stack image due to high-speed imaging. To remove the redundant empty frames, a variance-threshold-based algorithm was developed to reduce the stack size by >90%.
- Single-cell segmentation and quantification: Single cells captured by DC feature a thin bright halo around the cell body. Conventional image processing algorithms were applied to locate the cell contour and assess the morphological and intensity features (i.e., deforation, area, mean intensity, etc.).
Please refer to the GitHub repository and project report (pdf file) for more details.