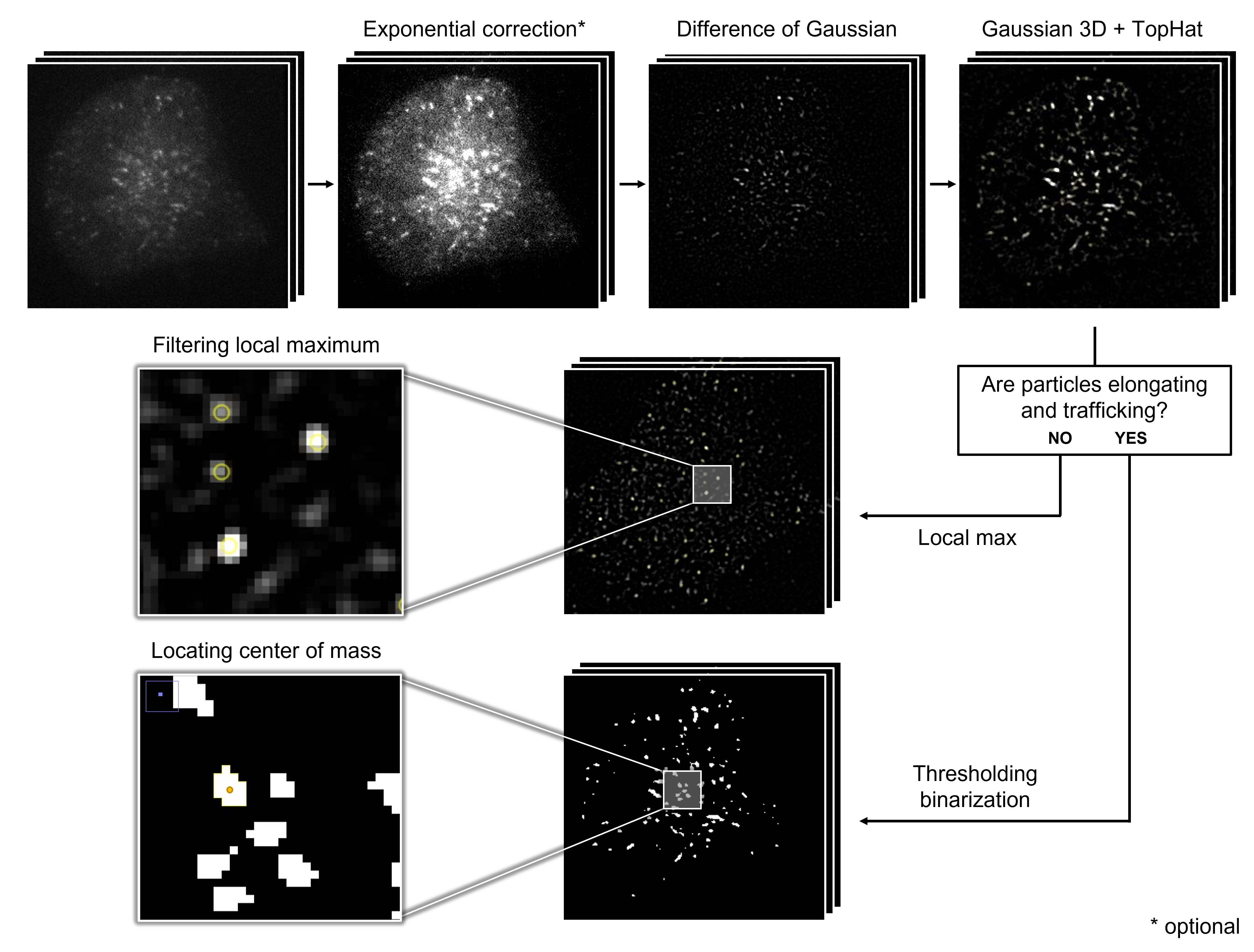
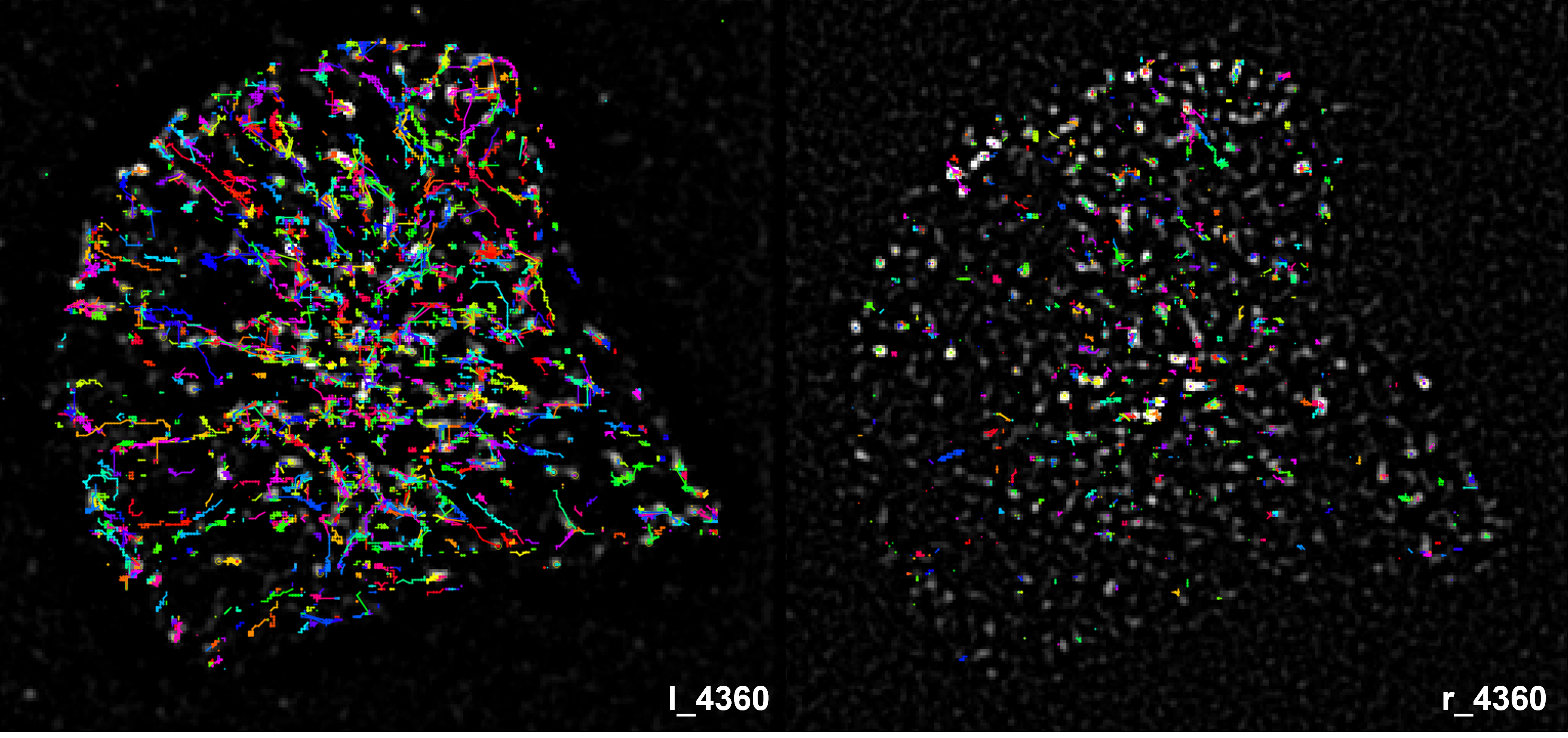
TIRF protein tracking and analysis
A bioimage processing workflow that detects, tracks and analyzes protein dynamics in TIRF microscopy images.
This project was done in group in Bioimage Informatics (2023 Spring) taught by Dr. Sage Daniel and Dr. Seitz Arne.
We built a particle tracking workflow for TIRF microscopy images. Specifically, I focused on the development of particle detection and trajectory. Please refer to the GitHub repository for more details.